From Tissue to Transcriptome: A Robust Pipeline for Single-Cell RNA-seq of Primary Porcine Hepatocytes
DOI:
https://doi.org/10.12775/TRVS.2025.003Keywords
Single-cell RNA sequencing (scRNA-seq), Porcine liver, Transcriptome integrityAbstract
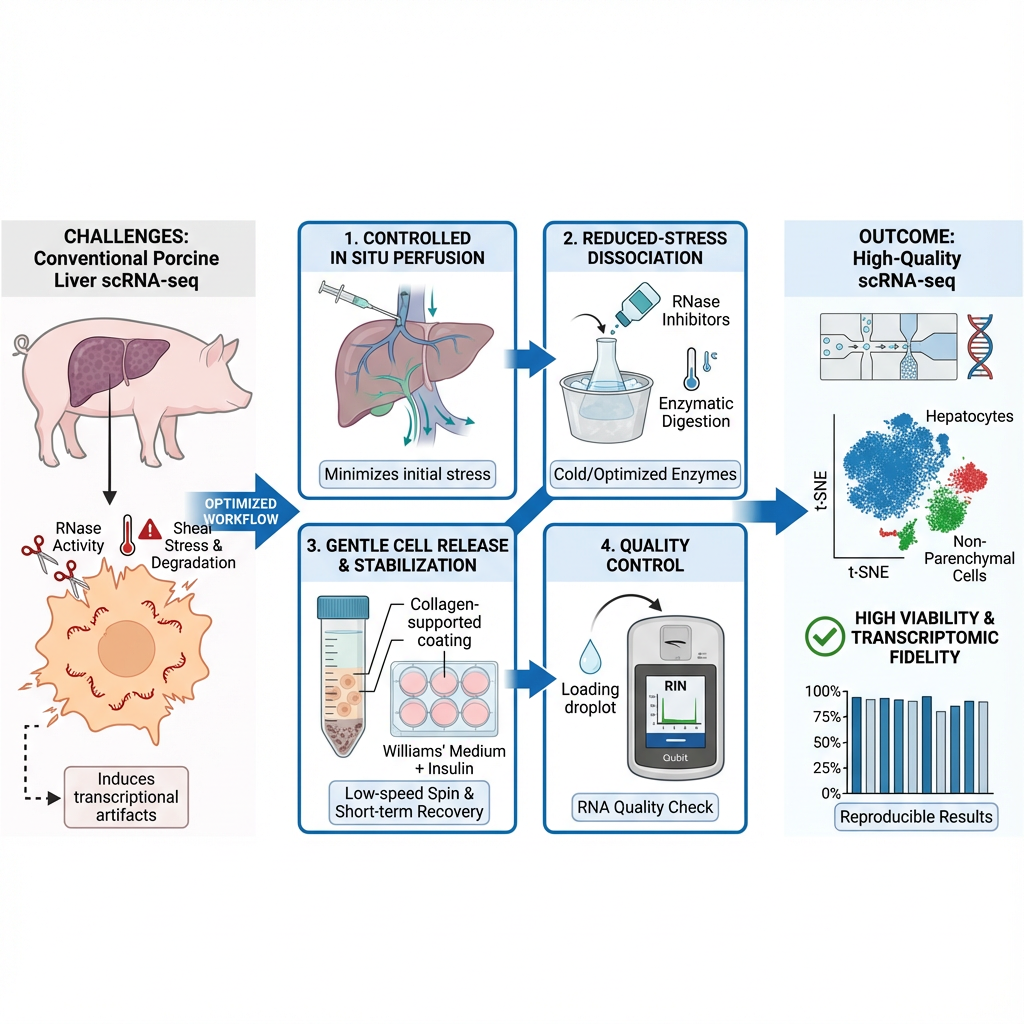
The technical execution of single-cell RNA sequencing (scRNA-seq) in liver tissue poses notable challenges due to the inherent fragility of hepatocytes, the distinct metabolic zonation, and the rapid activation of stress-response mechanisms during tissue dissociation. These complications are further exacerbated in porcine liver, where the large size of hepatocytes and elevated endogenous RNase activity resemble human hepatic tissue, yet are particularly vulnerable to mechanical and enzymatic stresses. Conventional scRNA-seq methodologies frequently yield diminished cell viability, RNA degradation, and skewed representation of hepatic subpopulations when applied to porcine models. We introduce a reproducible and species-optimized protocol for conducting scRNA-seq on porcine liver. This methodology incorporates cold-active enzymatic dissociation, low-shear mechanical manipulation, RNase suppression, and debris-minimized density separation to maintain cellular integrity and transcriptomic accuracy. A brief in vitro stabilization phase further improves cell viability and mitigates ambient RNA contamination, yielding single-cell suspensions amenable to droplet-based systems, such as the 10× Genomics v3.1 and v4 chemistries. Benchmarking outcomes reveal high viability rates (>80%), substantial transcript complexity, and consistent recovery of both parenchymal and non-parenchymal populations with diminished stress-response profiles. This workflow establishes a standardized approach for high-fidelity, cross-institutional scRNA-seq investigations and bolsters translational research applications in toxicology, pharmacokinetics, and hepatoprotective therapies.
Acknowledgement
This research is financed and supported by the scientific project NCN-OPUSLAP (UMO-2021/43/I/NZ9/02612) entitled: Multilevel molecular analysis of the hepatoprotective effect of medicinal herbs extracts in prevention of liver dysfunction caused by aflatoxin B1 in pig as an animal model (in-vivo), and hepatocyte cell culture analysis in human and pig (in-vitro).
References
1. Andrews, T. S., Atif, J., Liu, J. C., Perciani, C. T., Ma, X.-Z., Thoeni, C., Slyper, M., Eraslan, G., Segerstolpe, A., Manuel, J., Chung, S., Winter, E., Cirlan, I., Khuu, N., Fischer, S., Rozenblatt-Rosen, O., Regev, A., McGilvray, I. D., Bader, G. D., & MacParland, S. A. (2022). Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatology Communications, 6(4), 821–840. https://doi.org/10.1002/hep4.1854
2. Atif, J., Thoeni, C., Bader, G. D., McGilvray, I. D., & MacParland, S. A. (2022). Unraveling the complexity of liver disease one cell at a time. Seminars in Liver Disease, 42(3), 250–270. https://doi.org/10.1055/s-0042-1755272
3. Chałaśkiewicz, K., Kępka-Borkowska, K., Starzyński, R. R., Ogłuszka, M., Borkowski, M., Poławska, E., Lepczyński, A., Lichwiarska, E., Sultana, S., Kalra, G., Purohit, N., Pareek, C. S., & Pierzchała, M. (2025). Impact of aflatoxins on the digestive, immune, and nervous systems: The role of microbiota and probiotics in toxicity protection. International Journal of Molecular Sciences, 26(17), 8258. https://doi.org/10.3390/ijms26178258
4. Charni-Natan, M., & Goldstein, I. (2020). Protocol for primary mouse hepatocyte isolation. STAR Protocols, 1(2), 100086. https://doi.org/10.1016/j.xpro.2020.100086
5. Chen, L., Tong, X., Wu, Y., Liu, C., Tang, C., Qi, X., Kong, F., Li, M., Jin, L., & Zeng, B. (2025). A dataset of single-cell transcriptomic atlas of Bama pig and potential marker genes across seven tissues. BMC Genomic Data, 26(1), 16. https://doi.org/10.1186/s12863-025-01308-3
6. Cheng, S., Zou, Y., Zhang, M., Bai, S., Tao, K., Wu, J., Shi, Y., Wu, Y., Lu, Y., He, K., Sun, P., Su, X., Hou, S., & Han, B. (2023). Single-cell RNA sequencing reveals the heterogeneity and intercellular communication of hepatic stellate cells and macrophages during liver fibrosis. MedComm, 4(5), e378. https://doi.org/10.1002/mco2.378
7. Chu, A. L., Schilling, J. D., King, K. R., & Feldstein, A. E. (2021). The power of single-cell analysis for the study of liver pathobiology. Hepatology (Baltimore, Md.), 73(1), 437–448. https://doi.org/10.1002/hep.31485
8. Denisenko, E., Guo, B. B., Jones, M., Hou, R., de Kock, L., Lassmann, T., Poppe, D., Clément, O., Simmons, R. K., Lister, R., & Forrest, A. R. R. (2020). Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biology, 21(1), 130. https://doi.org/10.1186/s13059-020-02048-6
9. Dexter, L. J. (2012). The development of human primary hepatocyte model for investigating the pathogenesis of the hepatitis C virus [PhD, University of Nottingham]. https://eprints.nottingham.ac.uk/12510/
10. Esteves, F., Rueff, J., & Kranendonk, M. (2021). The central role of Cytochrome P450 in xenobiotic metabolism-A brief review on a fascinating enzyme family. Journal of Xenobiotics, 11(3), 94–114. https://doi.org/10.3390/jox11030007
11. Fujimoto, M., & Tanaka, T. (2025). Advancing liver metabolic zonation with single-cell and spatial omics. Endocrine Journal, 72(10), 1069–1078. https://doi.org/10.1507/endocrj.EJ25-0140
12. Genshaft, A. S., Subudhi, S., Keo, A., Sanchez Vasquez, J. D., Conceição-Neto, N., Mahamed, D., Boeijen, L. L., Alatrakchi, N., Oetheimer, C., Vilme, M., Drake, R., Fleming, I., Tran, N., Tzouanas, C., Joseph-Chazan, J., Arreola Villanueva, M., van de Werken, H. J. G., van Oord, G. W., Groothuismink, Z. M. A., … Gehring, A. J. (2023). Single-cell RNA sequencing of liver fine-needle aspirates captures immune diversity in the blood and liver in chronic hepatitis B patients. Hepatology (Baltimore, Md.), 78(5), 1525–1541. https://doi.org/10.1097/HEP.0000000000000438
13. He, J., Deng, C., Krall, L., & Shan, Z. (2023). ScRNA-seq and ST-seq in liver research. Cell Regeneration (London, England), 12(1), 11. https://doi.org/10.1186/s13619-022-00152-5
14. He, L., Lu, A., Qin, L., Zhang, Q., Ling, H., Tan, D., & He, Y. (2021). Application of single-cell RNA sequencing technology in liver diseases: a narrative review. Annals of Translational Medicine, 9(20), 1598. https://doi.org/10.21037/atm-21-4824
15. Jankelow, A., Almeida-Porada, G., Atala, A., Sawyer, S. W., & Porada, C. D. (2025). Recent advancements in tissue dissociation techniques for cell manufacturing single-cell analysis and downstream processing. Stem Cells Translational Medicine, 14(11). https://doi.org/10.1093/stcltm/szaf055
16. Junatas, K. L., Tonar, Z., Kubíková, T., Liška, V., Pálek, R., Mik, P., Králíčková, M., & Witter, K. (2017). Stereological analysis of size and density of hepatocytes in the porcine liver. Journal of Anatomy, 230(4), 575–588. https://doi.org/10.1111/joa.12585
17. Jung, Y., Zhao, M., & Svensson, K. J. (2020). Isolation, culture, and functional analysis of hepatocytes from mice with fatty liver disease. STAR Protocols, 1(3), 100222. https://doi.org/10.1016/j.xpro.2020.100222
18. Kegel, V., Deharde, D., Pfeiffer, E., Zeilinger, K., Seehofer, D., & Damm, G. (2016). Protocol for isolation of primary human hepatocytes and corresponding major populations of non-parenchymal liver cells. Journal of Visualized Experiments: JoVE, 109, e53069. https://doi.org/10.3791/53069
19. Kępka-Borkowska, K., Chałaśkiewicz, K., Ogłuszka, M., Borkowski, M., Lepczyński, A., Pareek, C. S., Starzyński, R. R., Lichwiarska, E., Sultana, S., Kalra, G., Purohit, N., Gralak, B., Poławska, E., & Pierzchała, M. (2025). Current approaches to aflatoxin B1 control in food and feed safety: Detection, inhibition, and mitigation. International Journal of Molecular Sciences, 26(13), 6534. https://doi.org/10.3390/ijms26136534
20. Kibitlewska, K., Asediya, V., Karpiesiuk, K., Czarnik, U., Lecewicz, M., Wysocki, P., Sharma, P., Otrocka-Domagała, I., Zielonka, Ł., Pomianowski, A., Okorski, A., Kalra, G., Sultana, S., Purohit, N., Lepczyński, A., Ożgo, M., Marynowska, M., Herosimczyk, A., Redlarska, E., … Kozera, W. (2026). Hepatoprotective potential of curcumin in the prevention of liver dysfunction in a porcine model. Nutrients, 18, 408. https://doi.org/10.3390/nu18030408
21. Li, W., Li, P., Li, N., Du, Y., Lü, S., Elad, D., & Long, M. (2021). Matrix stiffness and shear stresses modulate hepatocyte functions in a fibrotic liver sinusoidal model. American Journal of Physiology. Gastrointestinal and Liver Physiology, 320(3), G272–G282. https://doi.org/10.1152/ajpgi.00379.2019
22. Liang, S., & Acharya, K. R. (2016). Structural basis of substrate specificity in porcine RNase 4. The FEBS Journal, 283(5), 912–928. https://doi.org/10.1111/febs.13646
23. Lichwiarska, E., Ożgo, M., Pierzchała, M., Kępka-Borkowska, K., Chałaśkiewicz, K., Pareek, C. S., & Lepczyński, A. (2025). Impacts of Andrographis paniculata supplementation on health and productivity in monogastric farm animals: A comprehensive review. Animal Nutrition. https://doi.org/10.1016/j.aninu.2025.07.004
24. Liu, L., Besson-Girard, S., Ji, H., Gehring, K., Bulut, B., Kaya, T., Usifo, F., Simons, M., & Gokce, O. (2021). Dissociation of microdissected mouse brain tissue for artifact free single-cell RNA sequencing. STAR Protocols, 2(2), 100590. https://doi.org/10.1016/j.xpro.2021.100590
25. Lunney, J. K., Van Goor, A., Walker, K. E., Hailstock, T., Franklin, J., & Dai, C. (2021). Importance of the pig as a human biomedical model. Science Translational Medicine, 13(621), eabd5758. https://doi.org/10.1126/scitranslmed.abd5758
26. Machado, L., Relaix, F., & Mourikis, P. (2021). Stress relief: emerging methods to mitigate dissociation-induced artefacts. Trends in Cell Biology, 31(11), 888–897. https://doi.org/10.1016/j.tcb.2021.05.004
27. MacParland, S. A., Liu, J. C., Ma, X.-Z., Innes, B. T., Bartczak, A. M., Gage, B. K., Manuel, J., Khuu, N., Echeverri, J., Linares, I., Gupta, R., Cheng, M. L., Liu, L. Y., Camat, D., Chung, S. W., Seliga, R. K., Shao, Z., Lee, E., Ogawa, S., … McGilvray, I. D. (2018). Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature Communications, 9(1), 4383. https://doi.org/10.1038/s41467-018-06318-7
28. Mamanova, L., Miao, Z., Jinat, A., Ellis, P., Shirley, L., & Teichmann, S. A. (2021). High-throughput full-length single-cell RNA-seq automation. Nature Protocols, 16(6), 2886–2915. https://doi.org/10.1038/s41596-021-00523-3
29. N., Ponsuksili, S., Kozera, W., Karpiesiuk, K., Kępka-Borkowska, K., Chałaśkiewicz, K., Pierzchała, M., Taniguchi, H., Lepczyński, A., Ślaska, B., Asediya, V., Pareek, C. S., & Wimmers, K. (2026). Transcriptional insights into aflatoxin B1–induced hepatotoxicity and comparative effects of medicinal herbs in pigs. BMC Veterinary Research, 22, 53. https://doi.org/10.1186/s12917-025-05270-1
30. Pareek, C. S., Sachajko, M., Kalra, G., Sultana, S., Szostak, A., Chałaśkiewicz, K., Kępka-Borkowska, K., Poławska, E., Ogłuszka, M., Pierzchała, D., Starzyński, R., Taniguchi, H., Juszczuk-Kubiak, E., Lepczyński, A., Ślaska, B., Kozera, W., Czarnik, U., Wysocki, P., Kadarmideen, H. N., … Pierzchała, M. (2025). Identification of trait-associated microRNA modules in liver transcriptome of pig fed with PUFAs-enriched supplementary diet. Journal of Applied Genetics, 66(2), 389–407. https://doi.org/10.1007/s13353-024-00912-w
31. Payen, V. L., Lavergne, A., Alevra Sarika, N., Colonval, M., Karim, L., Deckers, M., Najimi, M., Coppieters, W., Charloteaux, B., Sokal, E. M., & El Taghdouini, A. (2021). Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Reports: Innovation in Hepatology, 3(3), 100278. https://doi.org/10.1016/j.jhepr.2021.100278
32. Pożarska, A., Karpiesiuk, K., Kozera, W., Czarnik, U., Dąbrowski, M., & Zielonka, Ł. (2024). AFB1 toxicity in human food and animal feed consumption: A review of experimental treatments and preventive measures. International Journal of Molecular Sciences, 25, 5305. https://doi.org/10.3390/ijms25105305
33. Rao, L., Cai, L., & Huang, L. (2023). Single-cell dynamics of liver development in postnatal pigs. Science Bulletin, 68(21), 2583–2597. https://doi.org/10.1016/j.scib.2023.09.021
34. Serras, A. S., Rodrigues, J. S., Cipriano, M., Rodrigues, A. V., Oliveira, N. G., & Miranda, J. P. (2021). A critical perspective on 3D liver models for drug metabolism and toxicology studies. Frontiers in Cell and Developmental Biology, 9, 626805. https://doi.org/10.3389/fcell.2021.626805
35. Sharma, P., Asediya, V., Kalra, G., Sultana, S., Purohit, N., Kibitlewska, K., Kozera, W., Czarnik, U., Karpiesiuk, K., Lecewicz, M., Wysocki, P., Lepczyński, A., Ożgo, M., Marynowska, M., Herosimczyk, A., Redlarska, E., Ślaska, B., Kowal, K., Tkaczyk-Wlizło, A., … Pierzchała, M. (2025). Hepatoprotective effect of silymarin herb in prevention of liver dysfunction using pig as animal model. Nutrients, 17(20), 3278. https://doi.org/10.3390/nu17203278
36. Van Melkebeke, L., Verbeek, J., Bihary, D., Boesch, M., Boeckx, B., Feio-Azevedo, R., Smets, L., Wallays, M., Claus, E., Bonne, L., Maleux, G., Govaere, O., Korf, H., Lambrechts, D., & van der Merwe, S. (2024). Comparison of the single-cell and single-nucleus hepatic myeloid landscape within decompensated cirrhosis patients. Frontiers in Immunology, 15, 1346520. https://doi.org/10.3389/fimmu.2024.1346520
37. Wang, S. (2022). Protocol to obtain high-quality single-cell RNA-sequencing data from mouse liver cells using centrifugation. STAR Protocols, 3(4), 101824. https://doi.org/10.1016/j.xpro.2022.101824
38. Wen, Y., & Ju, C. (2025). New insights into liver injury and regeneration from single-cell transcriptomics. EGastroenterology, 3(3), e100202. https://doi.org/10.1136/egastro-2025-100202
39. Xu, C., Fang, X., Xu, X., & Wei, X. (2024). Genetic engineering drives the breakthrough of pig models in liver disease research. Liver Research, 8(3), 131–140. https://doi.org/10.1016/j.livres.2024.09.003Nalpadan, A. A., Reyer, H., Oster, M., Trakooljul,
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Sharma P

This work is licensed under a Creative Commons Attribution-NoDerivatives 4.0 International License.
Title, logo and layout of TR in VS are reserved trademarks of TR in VR.
Stats
Number of views and downloads: 0
Number of citations: 0


